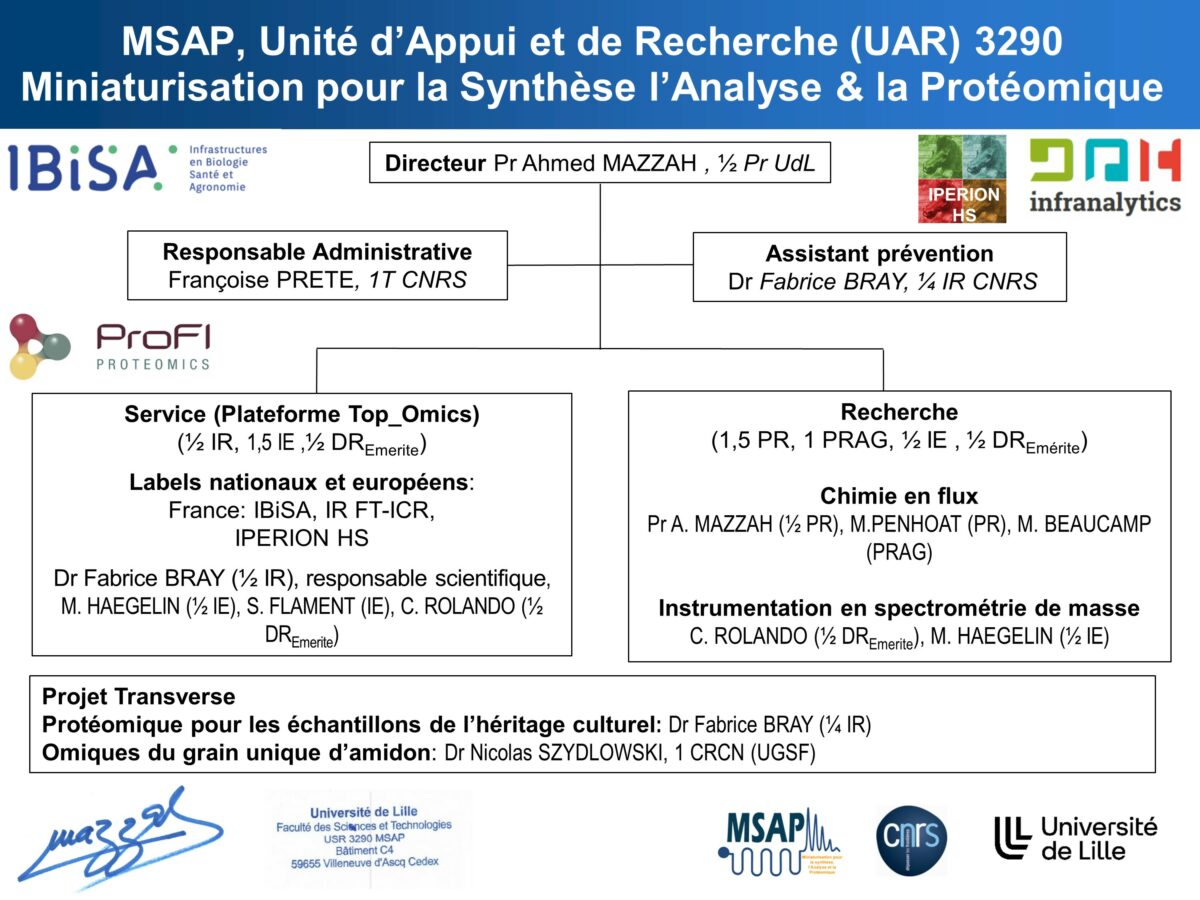
Founded in January 2010, UAR 3290 Miniaturisation pour la Synthèse, l’Analyse et la Protéomique (Miniaturization for Synthesis, Analysis and Proteomics), under the direction of Pr. Ahmed Mazzah, is a Support and Research Unit. UAR 3290 MSAP is accredited by IBiSA (Infrastructures en Biologie Santé et Agronomie), and is one of the sites of the national Infranalytics, and European IPERION HS and E-RIHS infrastructures. The unit’s proteomics service is integrated into MS4OMICS. This platform is divided into 2 parts, one located in the laboratory and the second in the SN3 building, attached to the PRISM U1192 Iserm laboratory. This platform has been awarded the ProFI Core label (national proteomics infrastructure).
The UAR 3290 is structured around three areas: mass spectrometry, separation techniques, organic synthesis and physical chemistry. These crafts are available in a service component focused on the mass spectrometry and proteomics (Bray Fabrice, IR) and a research component that includes: three scientific themes, strong interactions:
- Instrumental development (Christian Rolando, DR)
- Chemicals and flow chromatography (Mael Penhoat, MCF, Laetitia Chausset-Boissarie CR)
These different themes and service component lead us to work with most units of the Chevreul Institute. Structurally, the Unit is organized on a non-hierarchical model, each researcher or teacher researcher who support a themes and each engineer an analytical axes.
The theme proteomic analysis is a unifying theme for the other axis of the group. Indeed, it is:
- The main reason instrumental investment in mass spectrometry whose implementation is supported by our group (FT-MS high field MALDI TOF / TOF)
- The engine of most of the group’s analytical developments (new electrophoresis gels, specific fluorescent probes, surface chemistry and MALDI open capillary column type),
- At the heart of our collaborations biological purposes (especially the detection of biomarkers of different diseases related to aging)
- Is a new field for the analysis of complex systems such as archaeological residues, paleoproteomics.
- And causes miniaturization efforts have allowed us to address the organic reactivity studies in micro / nano reactors.
Proteomics is an integrating science. Indeed subjects with high added values to be able to involve from a biological sample and to identify and quantify proteins and post-translational modifications (phosphorylation, glycosylation). The main steps are:
- Protein extraction is a critical step, and that is the formulation in terms of chemistry to extract the expected proteins (particularly membrane proteins)
- Labeling them with specific reagents for quantification
- The separation involves a very complex set of chromatographic techniques (two-dimensional electrophoresis, exchange and size exclusion chromatography, immunoaffinity chromatography)
- Spectrometric analysis coupled with mass liquid chromatography nanoscale (flow is 200 nanoliters per minute). Typically 1000 proteins can be detected.
- The Bioinformatics treatment is a major bottleneck. Current analytical techniques often lead to the identification of hundreds of proteins which requires elaborate statistical treatments.